Publications
Nexomics Publications
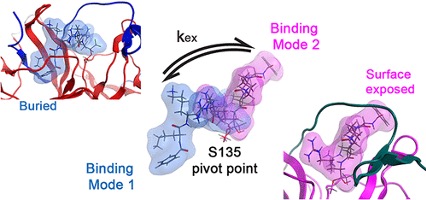
Gibbs AC, Steele R, Liu G, Tounge BA, Montelione GT. Inhibitor Bound Dengue NS2B-NS3pro Reveals Multiple Dynamic Binding Modes. Biochemistry. 2018 Mar 13;57(10):1591-1602. doi: 10.1021/acs.biochem.7b01127. Epub 2018 Feb 21. PubMed PMID: 29447443.
Yalpani N, Altier D, Barry J, Kassa A, Nowatzki TM, Sethi A, Zhao JZ, Diehn S, Crane V, Sandahl G, Guan R, Poland B, Perez Ortega C, Nelson ME, Xie W, Liu L, Wu G. An Alcaligenes strain emulates Bacillus thuringiensis producing a binary protein that kills corn rootworm through a mechanism similar to Cry34Ab1/Cry35Ab1. Sci Rep. 2017 Jun 8;7(1):3063. doi: 10.1038/s41598-017-03544-9. PMID: 28596570
Selected Publications of Nexomics Scientist
- Huang YJ, Zhang N, Bersch B, Fidelis K, Inouye M, Ishida Y, Kryshtafovych A, Kobayashi N, Kuroda Y, Liu G, LiWang A, Swapna GVT, Wu N, Yamazaki T, Montelione GT., Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2. Proteins. 2021 Dec;89(12):1959-1976. doi: 10.1002/prot.26246. Epub 2021 Oct 19. PMID: 34559429
- Koga N, Koga R, Liu G, Castellanos J, Montelione GT, Baker D., Role of backbone strain in de novo design of complex α/β protein structures. Nat Commun. 2021 Jun 24;12(1):3921. doi: 10.1038/s41467-021-24050-7. PMID: 34168113
- Aiyer S, Swapna GVT, Ma LC, Liu G, Hao J, Chalmers G, Jacobs BC, Montelione GT, Roth MJ.A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins. Structure. 2021 Feb 13:S0969-2126(21)00010-1. doi: 10.1016/j.str.2021.01.010. PMID: 33592170
- A. Cole, Nourhan S. Daigham, Gaohua Liu, Gaetano T. Montelione, Homayoun Valafar, REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEsCasey bioRxiv 2020.06.17.156638; doi: https://doi.org/10.1101/2020.06.17.156638.
- Sala D, Huang YJ, Cole CA, Snyder DA, Liu G, Ishida Y, Swapna GVT, Brock KP, Sander C, Fidelis K, Kryshtafovych A, Inouye M, Tejero R, Valafar H, Rosato A, Montelione GT. Protein structure prediction assisted with sparse NMR data in CASP13. Proteins. 2019 Dec;87(12):1315-1332. doi: 10.1002/prot.25837. PubMed PMID: 31603581; PubMed Central PMCID: PMC7213643.
- Koepnick B, Flatten J, Husain T, Ford A, Silva DA, Bick MJ, Bauer A, Liu G, Ishida Y, Boykov A, Estep RD, Kleinfelter S, Nørgård-Solano T, Wei L, Players F, Montelione GT, DiMaio F, Popović Z, Khatib F, Cooper S, Baker D. De novo protein design by citizen scientists. Nature. 2019 Jun;570(7761):390-394. doi: 10.1038/s41586-019-1274-4. Epub 2019 Jun 5. PubMed PMID: 31168091; PubMed Central PMCID: PMC6701466.
- Koepnick B, Flatten J, Husain T, Ford A, Silva DA, Bick MJ, Bauer A, Liu G, Ishida Y, Boykov A, Estep RD, Kleinfelter S, Nørgård-Solano T, Wei L, Players F, Montelione GT, DiMaio F, Popović Z, Khatib F, Cooper S, Baker D. De novo protein design by citizen scientists. Nature. 2019 Jun;570(7761):390-394. doi: 10.1038/s41586-019-1274-4. Epub 2019 Jun 5. PubMed PMID: 31168091; PubMed Central PMCID: PMC6701466.
Song, F.; Li, M.; Liu, G.; Swapna, G.V.T.; Daigham, N.S.; Xia, B.; Montelione, G.T.; Bunting, S.F. Biochemistry 2018, 57: 6568 – 6591. Antiparallel coiled-coil interactions mediate homodimerization of the DNA damage repair protein, PALB2. - Nie, Y.; Wang, S.; Xu, Y.; Luo, S.; Zhao, Y-L.; Xiao, R.; Montelione, G.; Hunt, J.; Szyperski, T. ACS Catalysis 2018, 8: 5145 – 5152. Enzyme engineering based on X-ray structures and kinetic profiling of substrate libraries: alcohol dehydrogenases for stereospecific synthesis of a broad range of chiral alcohols.
Zhang, M.; Yu, X.-W.; Swapna, G.V.T.; Liu, G.; Xiao, R.; Xu, Y.; Montelione, G.T. Biomol NMR Assign 2018, 12: 63 – 68. Backbone and Ile-δ 1, Leu, Val methyl 1H, 15N, and 13C, chemical shift assignments for Rhizopus chinensis lipase. - Marcos, E.; Basanta, B.; Chidyausiku, T.M.; Tang, Y.; Oberdorfer, G.; Liu, G.; Swapna, G.V.T.; Guan, R.; Silva, D.-A.; Dou, J.; Pereira, J.H.; Xiao, R.; Banumathi Sankaran, B.; Zwart, P.H.; Montelione, G.T.; Baker, D. Science 2017, 355: 201 – 206. Principles for designing proteins with cavities formed by curved β-sheets. (pdf) suppl. material
- Boël, G.; Letso, R.; Neely, H.; Price, W.N.; Wong, K.H.; Su, M.; Luff, J.; Valecha, M.; Everett, J.; Acton, T.B.; Xiao, R.; Montelione, G.T.; Aalberts, D.P.; Hunt, J.F. Nature 2016, 529: 358 – 363. Codon influence on protein expression in E. coli correlates to mRNA levels.
- Cai, K.; Liu, G.; Frederick, R.O.; Xiao, R.; Montelione, G.T.; Markley, J.L. Structure 2016, 24: 2080 – 2091. Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron-sulfur cluster biogenesis. (pdf) suppl. Material 1 (pdf) suppl. material 2 (pdf) PMC5166578
- Lin, Y-R.; Koga, N.; Tatsumi-Koga, R.; Liu, G.; Clouser, A.F.; Montelione, G.T.; Baker, D. Proc. Natl. Acad. Sci. U.S.A. 2015, 112: e5478 – 5485. Control over overall shape and size in de novo design proteins.
- Koga, N.; Tatsumi-Koga, R.; Liu, G.; Xiao, R.; Acton, T.B.; Montelione, G.T.; Baker, D. Nature 2012, 491: 222 – 227. Principles for designing ideal protein structures. (pdf) PMC3705962
- Thompson, J.; Sgourakis, N.G.; Liu, G.; Rossi, P.; Tang, Y.; Mills, J.; Szyperski, T.; Montelione, G.; Baker, D. Proc. Natl. Acad. Sci. USA 2012, 109: 9875 – 9880. Accurate protein structure modeling using sparse NMR data and homologous structure information. (pdf) suppl. material (pdf) PMC3382498
- Karanicolas, J.; Corn, J.E.; Chen, I; Joachimiak, L.A.; Dym, O.; Peck, S.H.; Albeck, S.; Unger, T.; Hu, W.; Liu, G.; Delbecq, S.; Montelione, G.T.; Spiegel, C.P.; Liu, D.R.; Baker, D. Molecular Cell 2011, 42: 250 – 260. A de novo protein binding pair by computational design and directed evolution. (pdf) suppl. material (pdf) PMC3102007
- Zhang, H.; Constantine, R.; Vorobiev, S.; Chen, Y.; Seetharaman, J.; Huang, Y.J.; Xiao, R.; Montelione, G.T.; Gerstner, C.D.; Davis, M.W.; Inana, G.; Whitby, F.G.; Jorgensen, E.M.; Hill, C.P.; Tong, L.; Baehr, W. Nature Neuroscience2011, 14: 874 – 880. UNC119 is required for G protein trafficking in sensory neurons. (pdf) suppl. material (pdf) PMC3178889
- Arbing, M.A.; Handelman, S.K.; Kuzin, A.P.; Verdon, G.; Wang, C.; Su, M.; Rothenbacher, F.P.; Abashidze, M.; Liu, M.; Hurley, J.M.; Xiao, R.; Acton, T.; Inouye, M.; Montelione, G.T.; Woychik, N.A.; Hunt, J.F. Structure 2010, 18: 996 – 1010. Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin-antitoxin systems.
- Liu, G.; Huang, Y.J.; Xiao, R.; Wang, D.; Acton, T.B.; Montelione, G.T. PROTEINS: Struct. Funct. Bioinformatics 2010, 78: 1326 – 1330. NMR structure F-actin binding domain of Arg/Ab12 from Homo sapiens. (pdf) suppl. material (pdf) PMC2821974
- Das, K.; Ma, L-C.; Xiao, R.; Radvansky, B.; Aramini, J.; Zhao, L.; Marklund, J.; Kuo, R-L.; Twu, K.Y.; Arnold, E.; Krug, R.M.; Montelione, G.T. Proc. Natl. Acad. Sci. U.S.A. 2008, 105: 13092 – 13097. Structural basis for suppression by influenza A virus of a host antiviral response. (pdf) suppl. material (pdf) PMC2522260
- Vila, J.; Aramini, J.; Rossi, P.; Kuzin, A.; Su, M.; Seetharaman, J.; Xiao, R.; Tong, L.; Montelione, G.T.; Scheraga, H. Proc. Natl. Acad. Sci. U.S.A. 2008, 105: 14389 – 14394. Quantum chemical 13Cα chemical shift calculations for protein NMR structure determination, refinement, and validation. (pdf) suppl. material (pdf) PMC2567219
- Shen, Y.; Lange, O.; Delaglio, F.; Rossi, P.; Aramini, J.M.; Liu, G.; Eletsky, A.; Wu, Y.; Singarapu, K.K.; Lemak, A.; Ignatchenko, A.; Arrowsmith, C.H.; Szyperski, T.; Montelione, G.T.; Baker, D.; Bax, A. Proc. Natl. Acad. Sci. U.S.A. 2008, 105: 4685 – 4690. Consistent blind protein structure generation from NMR chemical shift data. (pdf) suppl. material (pdf) PMC2290745
- Aramini, J.M.; Vorobiev, S.M.; Tuberty, L.M.; Janjua, H.; Campbell, E.T.; Seethraman, J.; Su, M.; Huang, Y.P.; Acton, T.B.; Xiao, R.; Tong, L.; Montelione, G.T. Structure 2015, 23: 1 – 12. The RAS-binding domain of human BRAF protein serine/threonine kinase exhibits allosteric conformational changes upon binding HRAS. (pdf) suppl. Material 1 (pdf) suppl. material 2 (pdf) PMC4963008
- Gräslund,S.; Nordlund, P.; Weigelt, J.; Bray, J.; Gileadi, O.; Knapp, S.; Oppermann, U.; Arrowsmith, C.; Hui, R.; Ming, J.; Park, H-w.; Savchenko, A.; Yee, A.; Edwards, A.; Vincentelli, R.; Cambillau, C.; Kim, R.; Kim, S-H.; Rao, Z.; Shi, Y.; Terwilliger, T.C.; Kim, C-Y.; Hung, L-W.; Waldo, G.S.; Peleg, Y.; Albeck, S.; Unger, T.; Dym, O.; Prilusky, J.; Sussman, J.L.; Stevens, R.C.; Lesley, S.A.; Wilson, I.A.; Joachimiak, A.; Collart, F.; Dementieva, I.; Donnelly, M.I.; Eschenfeldt, W.H.; Kim, Y.; Stols, L.; Wu, R.; Zhou, M.; Burley, S.K.; Emtage, J S.; Sauder, J.M.; Thompson, D.; Bain, K.; Luz, J.; Gheyi, T.; Zhang, F.; Atwell, S.; Almo, S.C.; Bonanno, J.B.; Fiser, A.; Swaminathan, S.; Studier, F.W.; Chance, M.R.; Sali, A.; Acton, T.B.; Xiao, R.; Zhao, L.; Ma, L-C.; Hunt, J.F.; Tong, L.; Cunningham, K.; Inouye, M.; Anderson, S.; Janjua, H.; Shastry, R.; Ho, C.K.; D.; Wang, H.; Jiang, M.; Montelione, G.T.; Stuart, D.I.; Owens, R.J.; Daenke, S.; Schütz, A.; Heinemann, U.; Yokoyama, S.; Büssow, K.; Gunsalus, K.C. Nature Methods 2008, 5: 135 – 146. Protein production and purification. (pdf) PMC3178102
Aramini, J.; Rossi, P.; Anklin, C.; Xiao, R.; Montelione, G.T. Nature Methods 2007, 4: 491 – 493. Microgram scale protein structure determination by NMR. (pdf) suppl. material (pdf) - Forouhar, F.; Anderson, J.L.; Mowat, C.G.; Vorobiev, S.M.; Hussain, A.; Abashidze, M.; Bruckmann, C.; Thackray, S.J.; Seetharaman, J.; Tucker, T.; Xiao, R.; Ma, L.; Zhao, L.; Acton, T.B.; Montelione, G.T.; Chapman. S.K.; Tong, L. Proc. Natl. Acad. Sci. U.S.A. 2007, 104: 473 – 478. Molecular insights into substrate recognition and catalysis by tryptophan 2,3- dioxygenase. (pdf) suppl. material (pdf)
- Liu, G.; Shen, Y.; Atreya, H.S.; Parish, D.; Shao, Y.; Sukumaran, D.K.; Xiao, R.; Yee, A.; Lemak, A.; Bhattacharya, A.; Acton, T.B.; Arrowsmith, C.H.; Montelione, G.T.; Szyperski, T. Proc. Natl. Acad. Sci. U.S.A. 2005, 102: 10487 – 10492. NMR data collection and analysis protocol for high-throughput protein structure determination. (pdf) suppl. material (pdf) PMC1180791
